Introduction
Subcellular ribosome assemblies are key components for synthesizing proteins inside the cell and translating genetic codes to the proteins. The ribosomes consist of two subunits and protein synthesis occurs when these subunits combine together to form a complete ribosome. The protein production capacity can be maximized by optimization of the ribosomal system. The number of ribosomes per cell and mass fraction of the active ribosome are among the key factors which control the production process. Ultracentrifugation has been employed to determine ribosome and subunits but the analysis is quite laborious and long.
Asymmetrical Flow Field-Flow Fractionation (AF4) has been successfully used as an alternative technique for separation and size characterization of ribosomes [1,2]. The separation in AF4 is based on exploiting differences in Diffusion Coefficients of sample constituents and the retention time of analytes can be converted to hydrodynamic diameter using the FFF retention theory. AF4 can be readily interfaced with other characterization techniques such as Multi Angle Light Scattering (MALS) for characterization of molar mass and root-mean-square radius (radius of gyration) of analytes.

In this application note two Escherichia Coli ribosome samples obtained from different growth procedures were analyzed by AF4-UV-MALS and compared with a control sample. The change in relative amount of the active ribosome compared to those of subunits may provide essential information to design optimal growth procedures.
AF4-UV-MALS Analysis of Escherichia Coli Ribosomes
Escherichia Coli ribosomes have an average molecular weight of 2.6 MDa with a sedimentation coefficient of 70S. They consist of two subunits of different size, the larger subunit with a sedimentation coefficient of 50S and a molecular weight of 1.6 - 1.8 MDa and the smaller one with a sedimentation coefficient of 30S and a molecular weight of 0.7 - 1.0 MDa. It is important to design conditions to have the majority of ribosomes actively involved in the protein synthesis process. This can be achieved by comparing the relative concentration of active ribosomes (70S) with those of subunits.
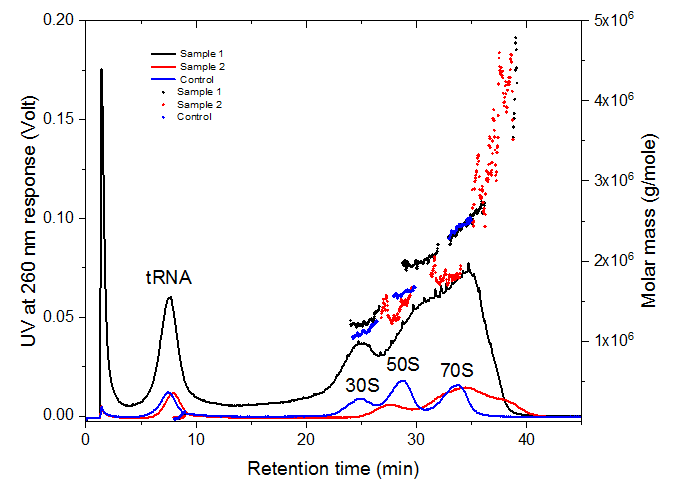
Figure 2 shows the results of the AF4-UV-MALS analysis of the control and two E. Coli ribosome samples. The blue line represents the UVbased fractogram of the control sample. The three peaks eluting at 24, 29 and 34 minutes correspond to 30S, 50S and 70S subunits and ribosome respectively.
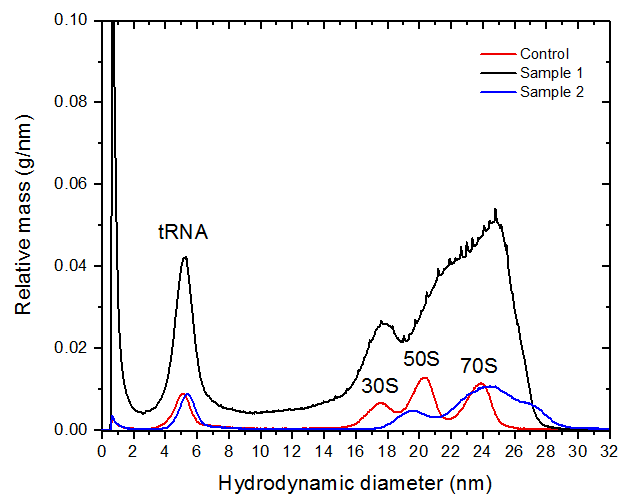
The identification of the peaks was confirmed by molar mass measured by MALS and the hydrodynamic particle diameter distributions obtained by FFF which are given in figure 3. The peak hydrodynamic diameters of 30S, 50S and 70S are measured as 18, 20 and 24 nm respectively. The values are in a good agreement with the literature values [1,2]. The mass fraction of the ribosomes in the samples was estimated from the UV peak maxima as 37 %, 66 % and 44 % for control, sample 1 and sample 2 respectively.
The peak eluting at 7 minutes corresponds to tRNA with an average molar mass of 30 kDa and a hydrodynamic diameter of 5 nm. Samples 1 and 2 exhibited some degree of ribosome aggregation which was detected by MALS. The molar mass of the aggregates was measured as 3 - 5 MDa for both samples.
Conclusion
Asymmetrical Flow Field-Flow Fractionation provides a high resolution separation methodology that can be used for size and mass characterization of ribosomes and their subunits in 40 minutes analysis time. The mass fraction of ribosomes can be estimated from peak maxima of the UV response. The method is also capable of separation and characterization of nonribosomal components and ribosome aggregates.
References
[1] Mikael N., Leif B., and Wahlund K.-G., Biotechnology and Bioengineering, 1997, 54(5), 461-467.
[2] Nilsson M., Birnbaum S., and Wahlund K.-G., Journal of Biomedical and Biophysical Methods, 1996, 33(1), 9-23.