Introduction
In Size Exclusion Chromatography (SEC), just like in Field-Flow Fractionation (FFF), the most preferred method to determine the molecular weight and molecular weight distribution of a polymer, biopolymer or protein/antibody sample is Multi Angle Static Light Scattering (MALS). MALS provides absolute molecular weight of a sample independent of molecular weight standards [1]. Furthermore, a MALS detector can also provide information about the molecular structure and branching conformation of a polymer or biopolymer sample. But sometimes there is only a concentration detector available. This is often a refractive index (RI) detector or a UV-vis detector, if the respective sample contains UV-active structures. In this case the method of conventional or relative calibration using well-defined standards can be used to determine the sample’s molecular weight and molecular weight distribution, respectively [2]. However, with this approach, information about the molecular structure of the sample is not directly accessible. In this application note, both MALS and relative calibration are compared with regard to their suitability for the reliable determination of the molecular weight of three branched dextran samples.
Relative vs. Absolute Molecular Weight Determination
In the following, a conventional calibration curve was established using a set of six linear, narrow Pullulan standards. Against this calibration curve three branched dextran samples with broad molecular weight distributions and different nominal molecular weights (Dextran 49 kDa (Dex 49), Dextran 148 kDa (Dex 148) and Dextran 273 kDa (Dex 273)) were measured. The used SEC-system comprised a Refractive Index detector (PN3150 RI), which served as concentration detector and a Multi Angle Light Scattering detector with 21 angles (PN3621 MALS) for absolute molecular weight determination. Analysis was performed in PBS buffer.
Conventional / Relative Calibration
For the method of conventional or relative calibration the elution volume respectively the retention time needs to be translated into molecular weight information. Therefore, a calibration curve has to be generated from a set of narrow polymer, biopolymer or protein standards of known molecular weight. In terms of polymers and biopolymers only linear standards are commercially available, such as the Pullulan standards used in this study for example.
Therefore, in order to determine the molecular weight of the dextran samples a set of six linear pullulan standards between 48.8 kDa and 348 kDa (PSS, USA) were measured. Plotting the molecular weight vs the elution/retention time derived from RI detection leads to the calibration curve displayed in Figure 1. Typically a logarithmic scale is used for the molecular weight axis.

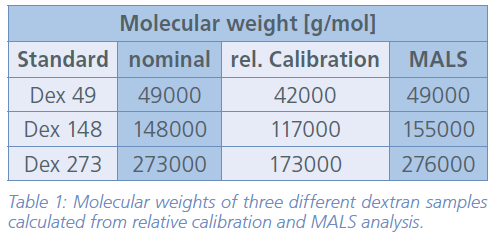
Once the relative calibration curve was set up the dextran samples were measured using the same conditions that were already established for the linear pullulan standards. From the elution time at each point of the corresponding peak the molecular weight of the dextrans was eventually calculated (Table 1).
Absolute Molecular Weight Determination with Static Light Scattering (MALS)
A MALS detector can be used to measure the absolute molecular weight of a polymer, biopolymer, protein or antibody sample. Here, the correlation between MALS-signal intensity and molecular weight is given by the Rayleigh light scattering equation:
MALS-Signal = KMALS x Conc x (dn/dc)2 x Mw
Mw is the weight average molecular weight, dn/dc is the refractive index increment and KMALS is the calibration constant. Instead of measuring several standards with narrow molecular weight distributions, the MALS detector can be calibrated with a single standard of known molecular weight, concentration and dn/dc value to determine the calibration constant. The absolute molecular weight of a sample can then be calculated directly from the MALS-signal while the RI detector measures the concentration of the sample at each point of the elution volume.
Results of Conventional or Relative Calibration vs. Light Scattering
To reveal the difference between relative and absolute molecular weight determination the results of the dextran measurements calculated from both relative calibration, and MALS detection were compared. A summary of the obtained results from both approaches is given in Table 1, clearly indicating an increasing deviation of the calculated molecular weights with increasing nominal molecular weight of the respective dextran samples (Figure 2). While MALS results are absolute and agree very well with the nominal values of the dextran samples here (Fig. 2, blue line), the deviation of the results obtained from relative calibration is striking, especially for the largest dextran sample (Dex 273). This observation can be explained by the lack of commercially available branched molecular weight standards. Hence, using a linear calibration curve for branched polymers with a higher molecular density compared to linear ones usually leads to an underestimation of the molecular weight of the respective polymers.

Conclusion
In this study, it was shown that due to the difference in structure for a branched sample compared to a calibration curve based on narrow, linear standards the relative results can differ significantly from the results obtained with a MALS detector. The same holds true if sample and standard do not have the same chemistry. It is therefore recommended to always use MALS for molecular weight determination especially of branched polymer samples as only MALS is able to measure the absolute molecular weight independent from the molecular structure or chemistry of the sample.
References
[1] P. Kratochvíl “Classical Light Scattering from Polymer Solutions” Elsevier, Amsterdam 1987, ISBN: 0-444-42890-9
[2] W. W. Yau, J. J. Kirkland and D. D. Bly “Modern Size-Exclusion Liquid Chromatography”, John Wiley & Sons, 2009, ISBN: 9780471201724